Cancer and brain development at single-cell resolution
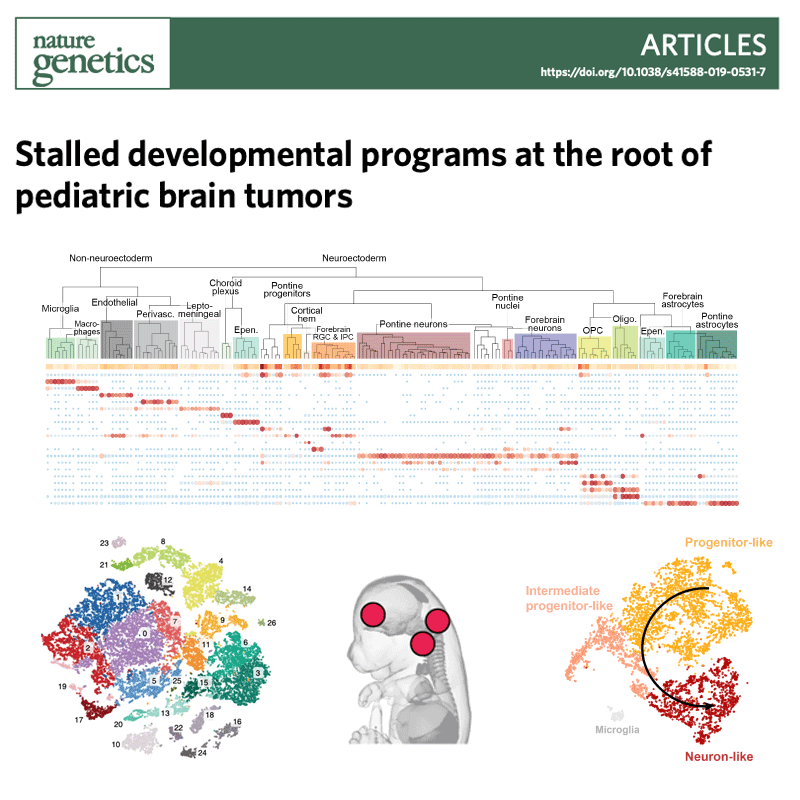
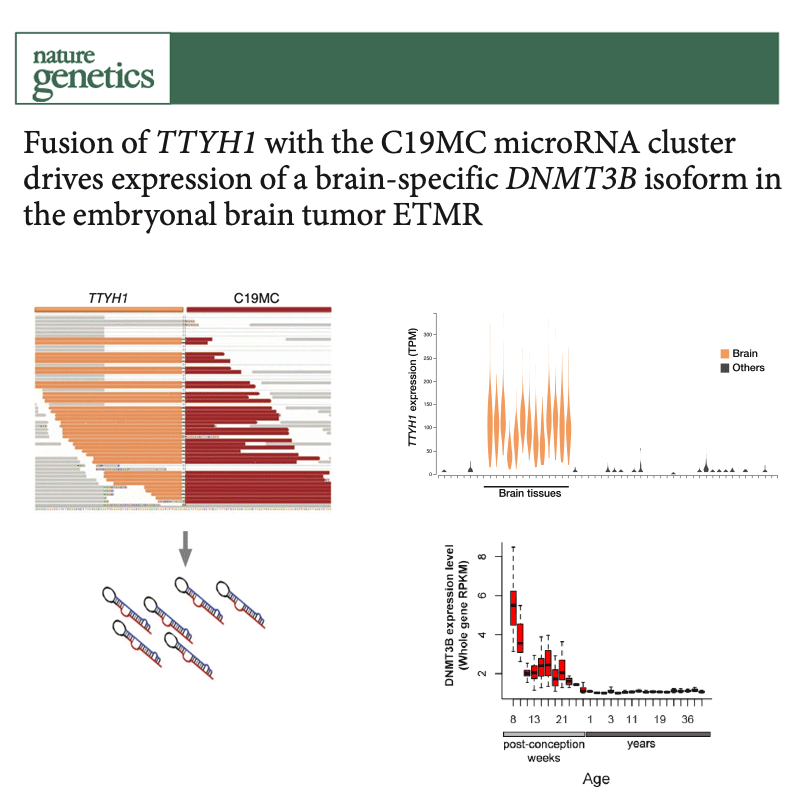
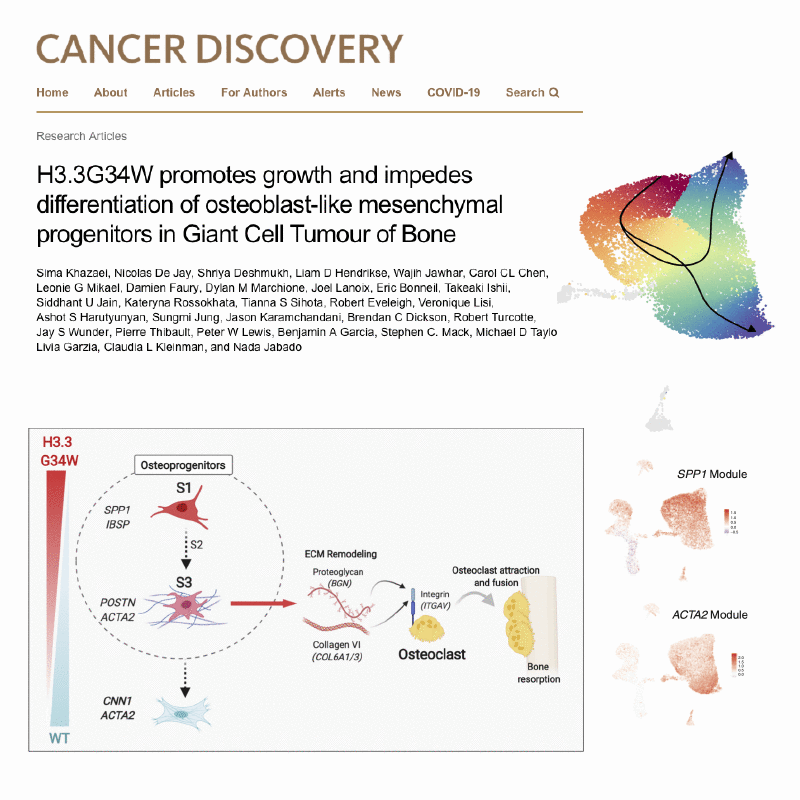
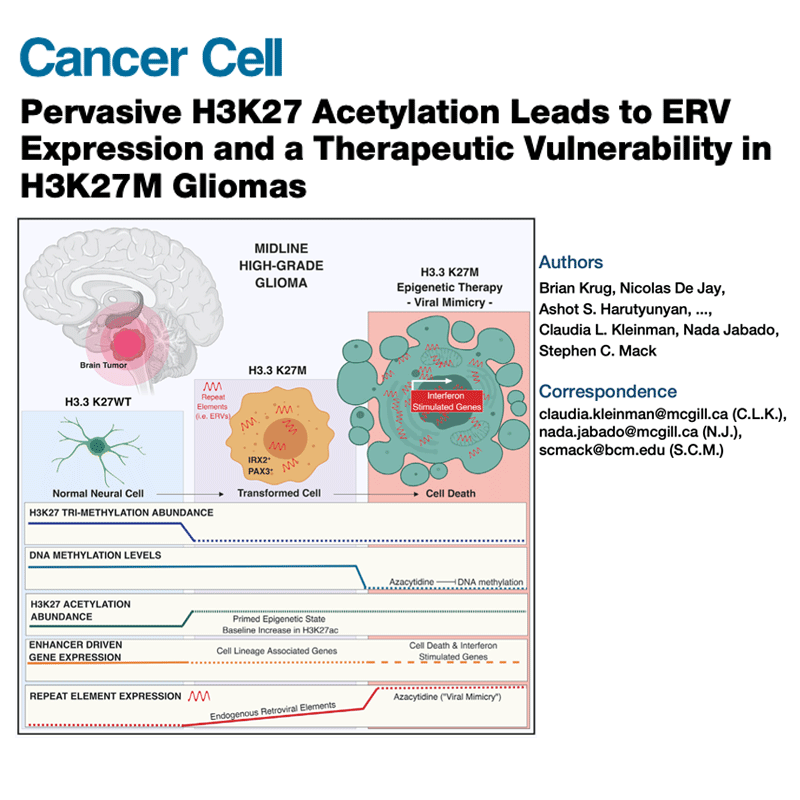
Several types of very aggressive and fatal pediatric brain tumors originate during brain development. The genetic event triggering the disease happens in the very earliest phases of cellular differentiation and, most likely, prenatally.
Our working model is that stalled development of progenitor cells is responsible for several childhood brain cancers.
Like Peter Pan in Never Land, cells are stuck in time, unable to progress towards a mature state. Instead of developing normally, the cells are arrested in a state that is permissive to malignant transformation.
But they retain key features of the original cells, which we use as fingerprints to pinpoint the cell of origin among the hundreds of different cell types present in the brain. For this, we are charting the normal developing brain at single-cell resolution, inspecting molecular profiles (transcriptome and epigenome) of hundreds of thousands of individual cells and integrating them with high-resolution, genome-wide tumor data, to define when and where brain tumors originate.
Resources from the lab:
Selected publications:
Read more in our Nature Cancer community blog postWatch a talk discussing our work on H3G34-mutant gliomas, by the co-first authors of the paper:
Integrative genomics
A major focus of the lab is on defining molecular mechanisms underlying cancer initiation, progression and therapeutic resistance. We take an integrative approach, leveraging multiple disease models (patient samples, cell lines, mouse models), and acquiring multimodal, genome-wide data.
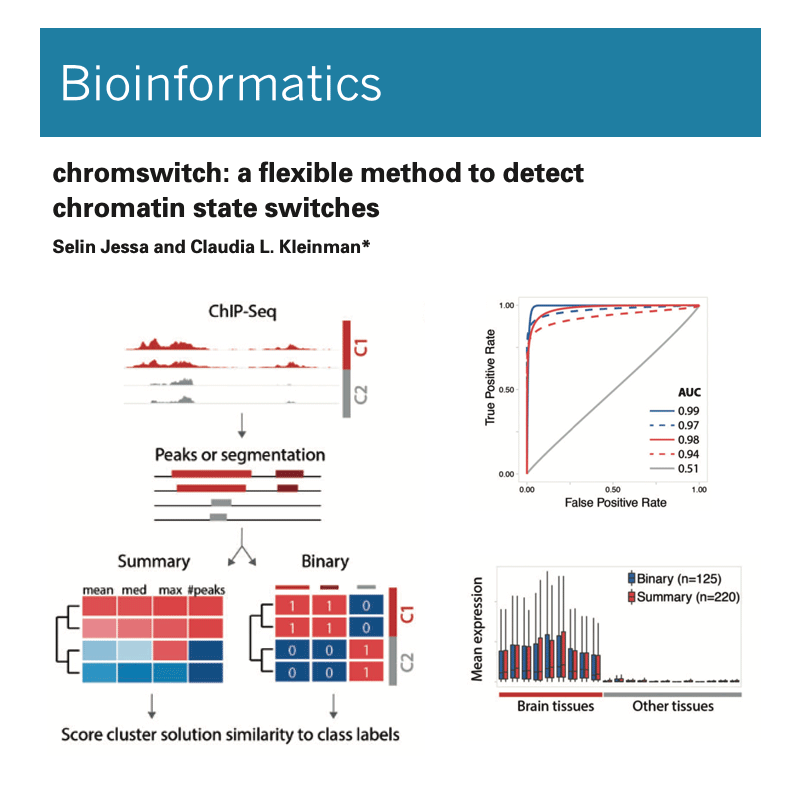
Integration of these genome-wide, mutimodal datasets requires developing novel computational strategies.
Selected publications:
Gene expression and RNA processing
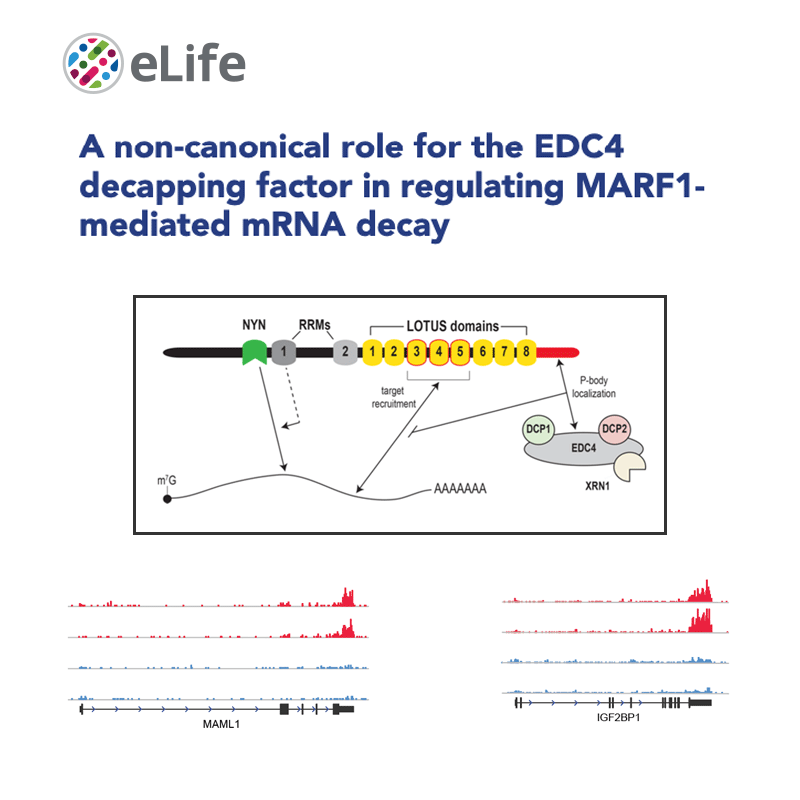
Gene expression regulation, both at the transcriptional and the post-transcriptional levels, is at the basis of essential biological processes, including cell identity specification, response to the environment and, of course, disease.
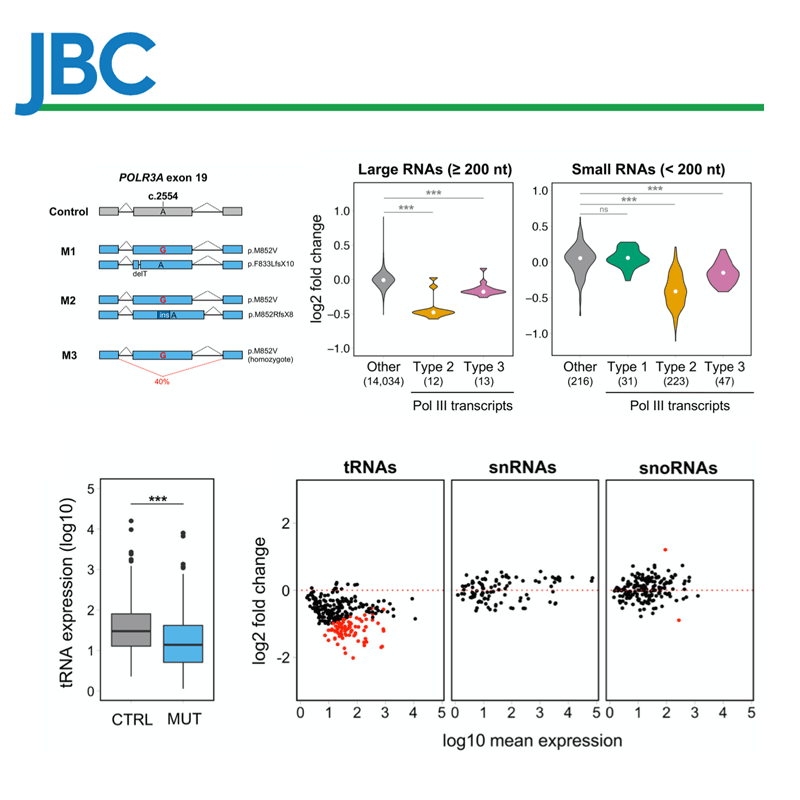
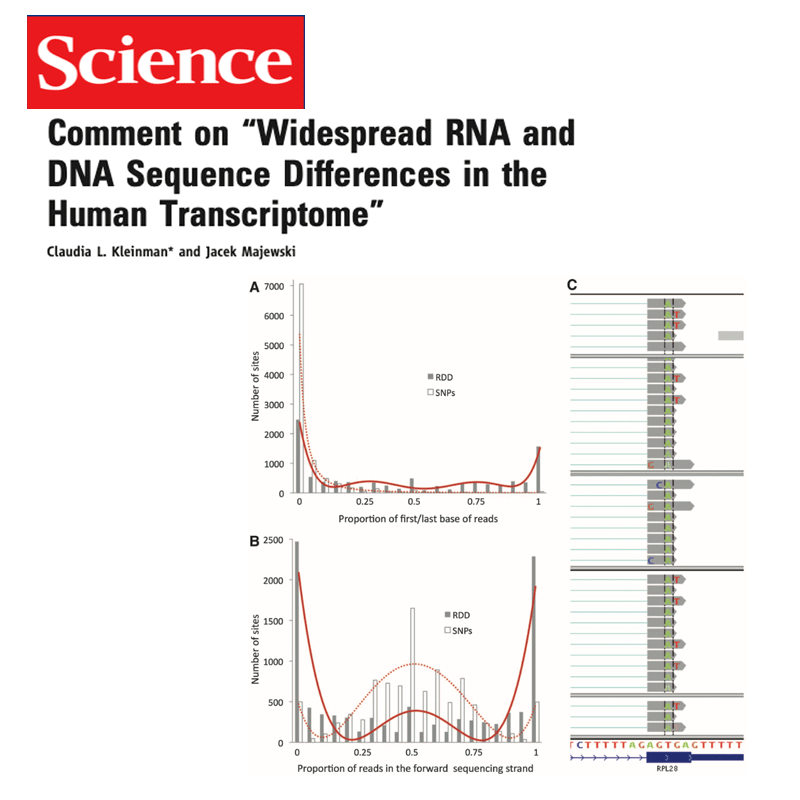
We have implemented methods for detecting RNA editing and other RNA chemical modifications, alternative splicing, non-coding RNA regulation and integration with epigenomic data.